
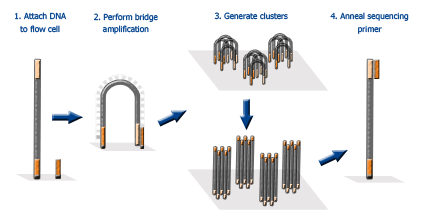
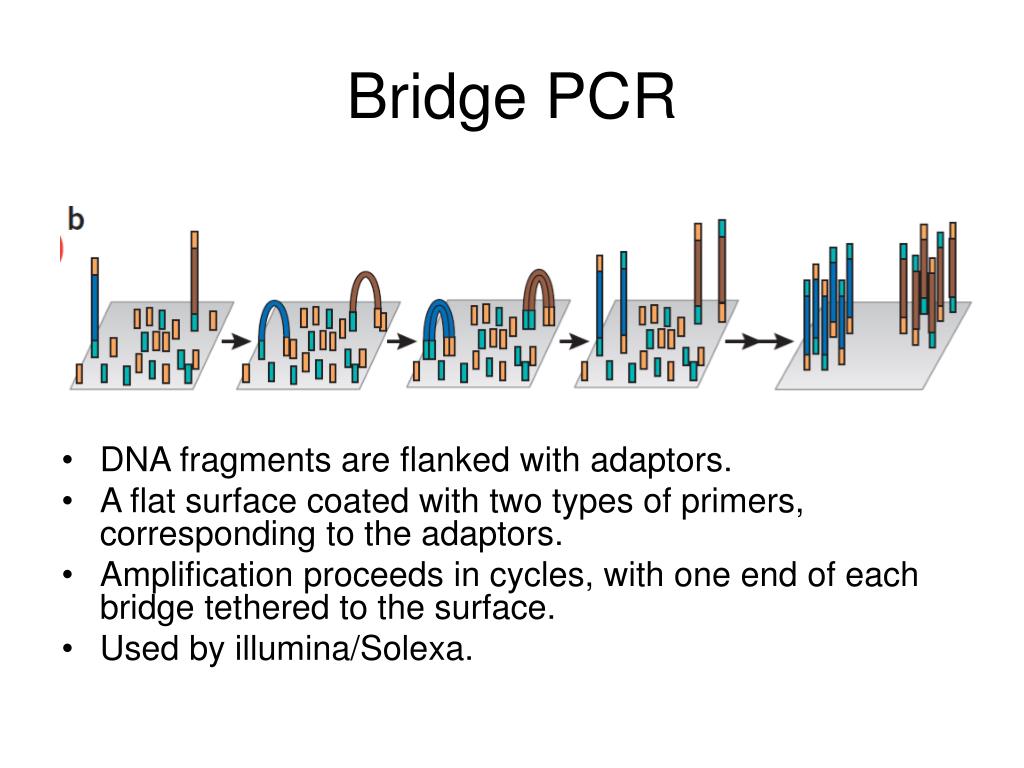
A plethora of genome sequencing projects that took many years with Sanger sequencing methods could now be completed within hours using NGS. The main difference between Sanger sequencing and 2G NGS stems from sequencing volume, with NGS allowing the processing of millions of reactions in parallel, resulting in high-throughput, higher sensitivity, speed and reduced cost. The bases of each fragment are then identified by their emitted signals. 11, 12 In 2G NGS, the genetic material (DNA or RNA) is fragmented, to which oligonucleotides of known sequences are attached, through a step known as adapter ligation, enabling the fragments to interact with the chosen sequencing system. The key principles behind Sanger sequencing and 2G NGS share some similarities.

9 In 2005, the first commercially available NGS platform, or second generation (2G) as it has become, was introduced, able to amplify millions of copies of a particular DNA fragment in a massively paralleled way in contrast to Sanger sequencing. With these immense technological advances, the human genome project was completed in 2003. These chain-terminated oligonucleotides are then size separated using gel electrophoresis in early methods, or capillary tubes in later automated capillary sequencers and the DNA sequence is determined. 8 When the DNA polymerase incorporates a ddNTP, the extension ceases leading to the generation of numerous copies of the DNA sequence of all lengths spanning the amplified fragment. The chain-termination method, also known as Sanger sequencing, uses a DNA sequence of interest as a template for a PCR that adds modified nucleotides, called dideoxyribonucleotides (ddNTPs), to the DNA strand during the extension step. 6, 7 This was the beginning of a golden era for the development and refinement of sequencing platforms, including the pivotal capillary DNA sequencer. 5 By 1986, the first automated DNA sequencing method had been developed. 3, 4 Various research groups then began adapting these methods to sequence DNA with a breakthrough coming in 1977 by Fredrick Sanger and colleagues, developing the chain-termination method. 1, 2 However, the first molecule to be sequenced was actually RNA – tRNA – in 1965 by Robert Holley and RNA of bacteriophage MS2 later on. The structure of DNA was determined in 1953 by Watson and Crick based on the fundamental DNA crystallography and X-ray diffraction work of Rosalind Franklin. Next-generation sequencing key terms and abbreviations Our simple polydisperse droplet preparation and statistical framework can be extended to a variety of settings for the quantification of nucleic acids in complex samples.What's the difference between whole-exome vs whole-genome sequencing? We also report a multiplexed ddPCR assay and demonstrate proof-of-concept methods for rapid droplet preparation in multiple samples simultaneously. In this work, we show that these ddPCR assays can reduce overall assay time while still providing quantitative results. Additionally, this approach is compatible with a range of input sample volumes, extending the assay dynamic range beyond that of commercial ddPCR systems. The polydisperse droplet system allows for accurate quantification of droplet digital PCR (ddPCR) and reverse transcriptase droplet digital PCR (RT-ddPCR) that is comparable to commercially available systems such as BioRad's ddPCR.
To address these limitations and make this technology more applicable for a variety of settings, we have developed a statistical framework to apply to droplet PCR performed in polydisperse droplets prepared without any specialized equipment. Though impactful, these improvements have generally been restricted to centralized laboratories with trained personnel and expensive equipment. These individual reaction vessels lead to digitization of PCR enabling improved time to detection and direct quantification of nucleic acids without a standard curve, therefore simplifying assay analysis. Lab-on-a-chip applications have developed methods to partition single biomolecules, such as DNA and RNA, into picoliter-sized droplets. Nucleic acid amplification technology, such as polymerase chain reaction (PCR), has enabled highly sensitive and specific disease detection and quantification, leading to more accurate diagnosis and treatment regimens.


 0 kommentar(er)
0 kommentar(er)
